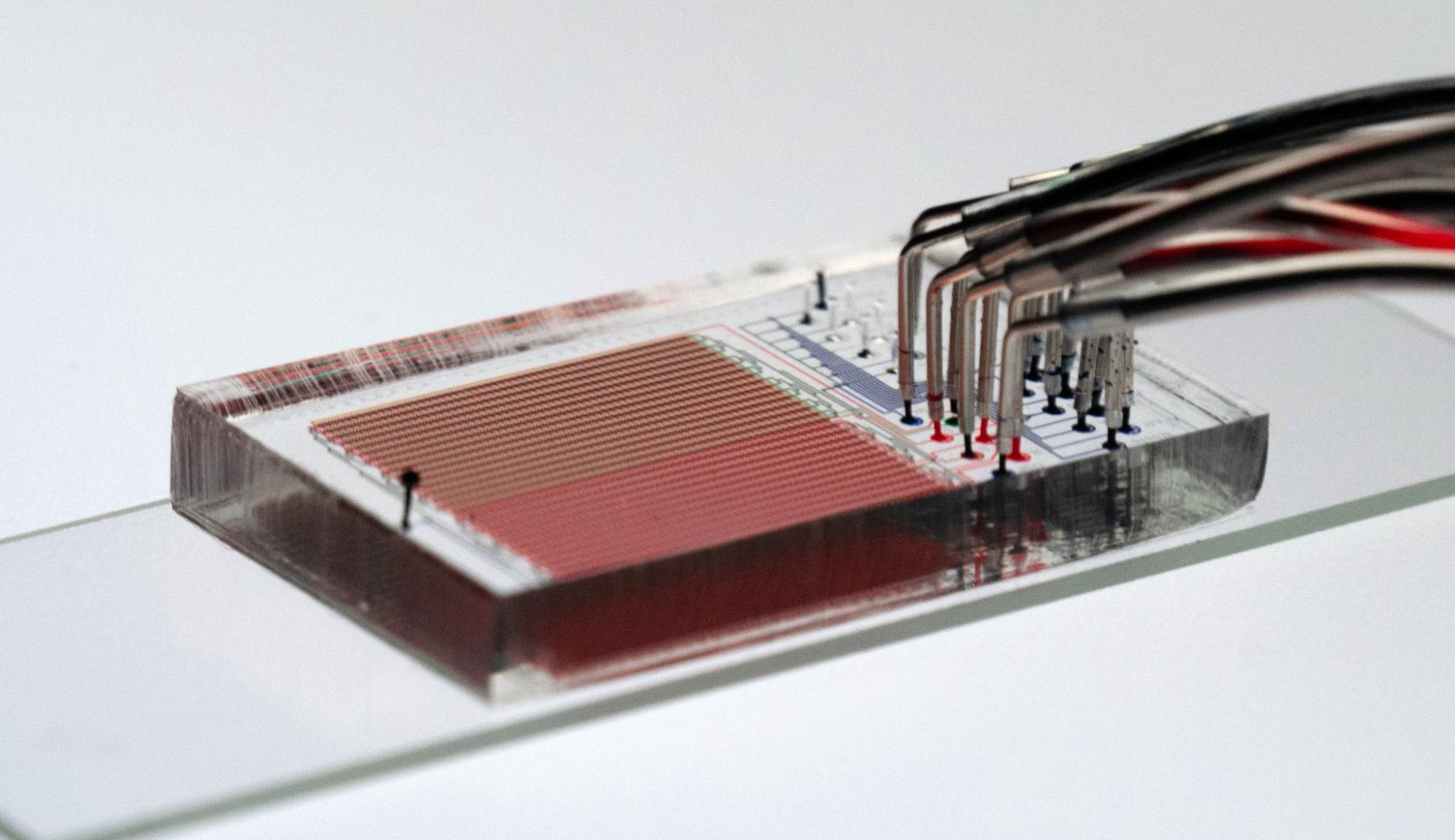
Polly Fordyce, who directs the Fordyce Lab, was awarded the Pioneer Award — a High Risk, High Reward grant from the National Institutes of Health (NIH) in October. The microfluidics approach to cancer immunotherapy research at the lab hopes to change and potentially personalize cancer treatments.
The lab received $700,000 per year in funding for the next five years on an ongoing project: “Using microfluidics to realize patient-specific anti-cancer immunotherapies.”
This biophysics-centered approach will leverage high-throughput microfluidics technologies to assess not only chemical binding, but also physical forces between immune cells and cancer cells. The approach hopes to identify the mechanisms through which the body’s immune response against cancer is activated.
“The ideas that led to this award are not just my ideas,” Fordyce said.
She praised the work from her group, including postdoctoral scholars and graduate students: “It is their work and their ideas that help us come up with new things.”
Fordyce also expressed gratitude for previous research that laid the groundwork for her current work.
She referenced contributions from bioengineering associate professor Stanley Qi, who was a key contributor to the invention of the CRISPR gene editing technique, molecular and cellular physiology professor Chris Garcia and chemical engineering associate professor Alex Dunn, as well as their respective labs.
“People have been thinking a lot about how we can harness a patient’s own immune system in order to fight cancer,” Fordyce said.
As Fordyce described, T-cells patrol the body for pathogens or cancerous cells by detecting proteins, or antigens, on antigen-presenting cells (APCs). Chemical and physical bonds form between an APC’s peptide-MHC (pMHC) complexes and a T-cell’s T-cell receptors (TCRs). T-cells become activated when they bind to foreign antigens, including neoantigens, which are the specific class of antigens displayed by cancerous cells. Once activated, T-cells kill the non-self cell.
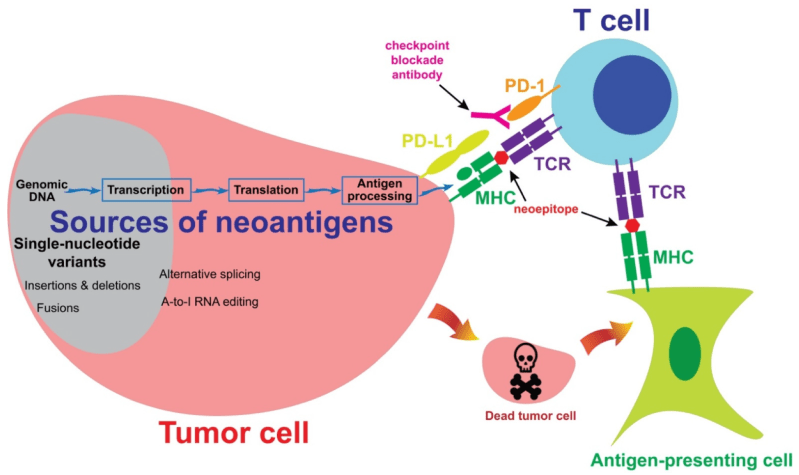
Current immunotherapy approaches screen millions of pMHC/TCR combinations for strong binding interactions between T-cells and antigen presenting cells. These interactions are then targeted with techniques such as T-cell transfusions and neoantigen vaccines to eliminate cancerous cells.
There are an intractably massive number of potential TCR/pMHC interactions, and only a select few may result in a potent yet targeted T-cell response against cancer, according to Matt DeJong, a chemical engineering Ph.D. student and one of the lead graduate student researchers in the Fordyce Lab.
“This molecular balancing act is truly a molecular feat, and we lack the data to understand and predict how T-cells specifically target sick cells in a sea of healthy cells,” DeJong said.
According to Fordyce, another challenge is that binding affinity, or how strongly a TCR chemically binds to a pMHC, does not necessarily predict T-cell activation. To approach this problem, the Fordyce Lab has taken a “very physical approach to understanding cancer biology,” Fordyce said.
In the grant abstract, the team wrote that the pMHC/TCR complexes formed between self-cells and T-cells form “slip” bonds that are more likely to break under force. Meanwhile, the pMHC/TCR complexes formed between T-cells and non-self cells form “catch” bonds that strengthen under force.
Catch bonds “are like those finger trap toys where you pull a bit harder and the bond gets stronger,” Fordyce said.
“If that is the clue telling us how T-cells recognize what is cancerous,” we need new tools to identify the molecular mechanisms behind that process, Fordyce said.
The Fordyce Lab is a microfluidics lab that specializes in developing new devices and platforms to miniaturize experiments from the scale of petri dishes and test tubes, to nanoliter and picoliter compartments.
This approach enables “lots of independent experiments all at once in something that’s about the size of a credit card,” said Dunn, who collaborates with the Fordyce Lab and co-advises DeJong with Fordyce.
In previous coordination with the Stanford Microfluidics Foundry, the lab developed a valved microfluidic device that allows for the production and purification of up to 1500 proteins in hours. “We are using this tool to ask which peptides are loaded efficiently into which MHC complexes, and which TCRs recognize these molecules to drive T-cell activation,” Fordyce said.
Fordyce said that the composition of pMHC complexes and proteins found on APCs are unique to each individual person’s immune system.
“The vision, which is well in the future, is that if you have a tumor with a specific mutation driving your specific cancer, we could make an mRNA vaccine to make that protein,” Fordyce said.
Alongside their existing microfluidics approaches, the lab plans to implement a higher throughput droplet-based technology to analyze tens of millions of T-cell/APC pairs.
“This approach amplifies the throughput massively compared to traditional plate-based assays,” DeJong said.
The team plans to use this droplet-based approach to sort the resulting pMHC/TCR combinations based on T-cell activation and possible “catch” bond formation.
“We can use flow cytometry machines to sort droplets on the basis of T-cell activation. That way, if a T-cell is activated, the droplet lights up,” Fordyce said.

Dunn emphasized the relevance of the Fordyce Lab’s research beyond personalized cancer treatments.
“This approach could enable all sorts of future strategies for engineering the human immune system,” he said.
Ecohing Dunn, Chaitain Khosla, a chemical engineering and chemistry professor unaffiliated with the research and grant, stressed the research’s importance.
“It’s great that one of Sarafan ChEM-H’s institute scholars is taking on this important problem of decoding T-cell recognition at scale. I can’t imagine a better colleague than Polly solving it,” he said.